Can Deep Learning Predict Race from Skin Histology Images? A New Warning for AI Fairness
2025-08-13
Citation: http://arxiv.org/pdf/2507.21912v2
Authors and Affiliations
- Shengjia Chen, Ruchika Verma, Jannes Jegminat, Eugenia Alleva, Thomas Fuchs, Gabriele Campanella, Kuan-lin Huang: Windreich Department of Artificial Intelligence and Human Health, Icahn School of Medicine at Mount Sinai, New York, USA, etc.
- Kevin Clare, Brandon Veremis: Department of Pathology, Molecular and Cell-Based Medicine, Mount Sinai Health System, New York, USA
Paper Summary
This paper, "Predicting patient self-reported race from skin histopathology images with deep learning," is a study focusing on a critical issue in the field of computational pathology[Note 1]: unintentional bias learning in AI models. The research examines whether a deep learning model can predict a patient's self-reported race from digital images of pathological tissue and clarifies what morphological "shortcuts"[Note 2] are used in making those predictions.
Research Objective: The main objective of this study is to verify whether a deep learning model can identify race from skin histology images and, if so, to identify the biological features that serve as the basis for its judgment. This aims to gain insights for prospectively evaluating and mitigating the risk of medical AI unintentionally disadvantaging specific populations due to hidden biases when applied in clinical settings.
Research Background: While AI, particularly deep learning, has shown remarkable success in disease detection and prognosis prediction, it has also been pointed out that it can learn biases present in training data, potentially exacerbating or amplifying existing healthcare disparities. In medical imaging, such as X-rays, it has been reported that AI can predict race with high accuracy from features imperceptible to experts, sparking significant debate. However, it remained largely unknown whether similar predictions were possible with histopathology images, which observe structures at the cellular level. Skin is particularly interesting because its appearance (skin color) is associated with race, but this difference becomes less obvious in stained tissue specimens, making it a compelling question what cues an AI might use.
Highlights of the Proposed Method: The most crucial aspect of the approach proposed in this study is the use of an AI model equipped with an attention mechanism[Note 3] to visualize "where" in the image the model is focusing when predicting race. As a result, they discovered that the AI uses a specific tissue structure, the "epidermis," as a strong cue to predict race. This is a groundbreaking achievement that concretely demonstrates the risk of AI learning biological features correlated with race as a "shortcut," rather than the disease itself.
Related Work
Review of Prior Research
- Race Prediction in Medical Imaging: Previous studies have shown that deep learning models can predict self-reported race from chest X-rays and MRI images with surprisingly high accuracy (AUC > 0.9). This ability is presumed to be based on subtle information that is not recognizable by human experts.
- Bias in Histopathology Images: In the field of pathology, it has been noted that differences in staining methods and scanner characteristics across research facilities (site signatures) can correlate with ethnicity, potentially affecting model performance and fairness. It has also been reported that models trained for diagnostic tasks a can unintentionally learn race information.
Differences Between Prior Research and This Study
While previous research primarily focused on race prediction in radiological images or technical biases in pathology images, this study is unique in that it narrows its focus to the specific field of dermatopathology and attempts to identify the biological and morphological cues (shortcuts) that enable race prediction. It does not simply report that "the model could predict race" but delves deeper into the basis of its judgment through attention analysis and UMAP visualization.
Novelty and Contributions
Novelty
This study offers a new perspective compared to existing research in the following ways:
- Novelty 1: It is one of the first studies to systematically investigate the ability of an AI model to predict self-reported race using skin histopathology images and to quantitatively evaluate its performance.
- Novelty 2: By combining attention analysis with morphological verification by pathologists, it identified a specific biological shortcut, the "epidermis," as the basis for prediction.
- Novelty 3: To clarify the impact of confounding factors[Note 4] such as disease distribution on prediction performance, it devised a tiered data curation strategy and rigorously compared its effects.
Contributions
This study makes the following academic and practical contributions:
- Contribution 1: It concretely demonstrates the risk that AI models in computational pathology can learn demographic information unrelated to disease and use it as a "shortcut" for prediction.
- Contribution 2: It proves that to develop fair medical AI, it is essential not only to pursue model performance but also to implement careful data management and techniques for interpreting the model's decision-making process.
- Contribution 3: It serves as a warning to the AI development community to be mindful of biases that models may unintentionally learn, providing an important foundation for future bias mitigation research.
Details of the Proposed Method
Method Overview
This paper uses a pipeline that combines a Foundation Model (FM) and Attention-based Multiple Instance Learning (AB-MIL) to classify race from skin histology images.
- A Foundation Model (FM)[Note 5] is used to divide a whole-slide image (WSI) of tissue into numerous small tile images and extract features from each tile. In this study, four different pre-trained models—SP22M, UNI, GigaPath, and Virchow—were tested.
- An Attention-based Multiple Instance Learning (AB-MIL)[Note 3] model is used to integrate the features from all tiles and predict race. The attention mechanism calculates the contribution (attention score) of each tile that was important for the prediction.
Method Architecture
The proposed method consists of the following steps:
- Data Preparation and Curation: Skin histopathology slides were collected from a patient population with diverse racial compositions at the Mount Sinai Health System. For the experiment, three datasets were created to control for confounding factors: unadjusted, disease-balanced, and based on strict ICD code criteria.
- Feature Extraction: Each slide is tiled at 20x magnification, and feature vectors for each tile are generated using four different FMs.
- Classification and Attention Score Calculation: The AB-MIL model is trained to predict the race label for the entire slide. Simultaneously, it calculates an attention score for each tile, indicating how much it contributed to the prediction.
- Interpretation and Analysis: UMAP[Note 6] is used to visualize high-attention tiles, and pathologists identify their morphological features (e.g., epidermis, inflammation). Furthermore, an ablation study is conducted to verify the importance of the epidermal regions.
Evaluation and Discussion
Evaluation Method
- Analysis Method: An AI model was trained on three data curation strategies (Exp1, Exp2, Exp3), and the changes in its performance were comparatively analyzed.
- Dataset: 5,266 skin tissue slides obtained from 2,471 patients at the Mount Sinai Health System. The dataset includes White, Black, Hispanic/Latino, Asian, and Other racial groups.
- Evaluation Metrics: AUC (Area Under the Curve)[Note 7] and Balanced Accuracy were used to evaluate the model's classification performance.
Research Findings
The results revealed that AI can predict race and that this prediction is strongly influenced by biases present in the data.
| Experiment | Encoder | White | Black | Hispanic | Asian | Other | Overall AUC | Overall Acc. |
|---|---|---|---|---|---|---|---|---|
| Exp1 | UNI | 0.797 | 0.791 | 0.607 | 0.791 | 0.603 | 0.718 | 0.400 |
| (Unadjusted) | Avg | 0.789 | 0.770 | 0.596 | 0.795 | 0.563 | 0.702 | 0.394 |
| Exp2 | UNI | 0.760 | 0.773 | 0.560 | 0.715 | 0.569 | 0.676 | 0.380 |
| (Disease-balanced) | Avg | 0.742 | 0.754 | 0.560 | 0.724 | 0.574 | 0.671 | 0.364 |
| Exp3 | UNI | 0.819 | 0.766 | 0.654 | 0.556 | 0.594 | 0.678 | 0.296 |
| (Strict ICD code) | Avg | 0.799 | 0.762 | 0.640 | 0.570 | 0.543 | 0.663 | 0.302 |
Table 1: Model performance across three dataset curation strategies. AUC is calculated using a One-vs-Rest approach.
-
Finding 1 (Impact of Bias):
- Exp1 (Unadjusted data): Showed the highest overall performance, but this was due to a skewed disease distribution (bias) where cases of "hemorrhoids" were unusually high (61%) in the data for Asian patients. The model was predicting based on a specific disease rather than race itself.
- Exp2 & Exp3 (After data adjustment): Correcting for the disease skew significantly reduced the prediction performance, especially for the Asian group. On the other hand, for the White (AUC 0.799) and Black (AUC 0.762) groups, high prediction performance was maintained even after removing the bias. This suggests the existence of race-specific morphological features separate from the disease in these groups.
-
Finding 2 (Identification of Shortcuts):
-
UMAP Visualization: When the regions the model focused on during prediction (high-attention regions) were visualized with UMAP, it was found that attention was concentrated in regions corresponding to the "epidermis," especially for the White and Black groups.
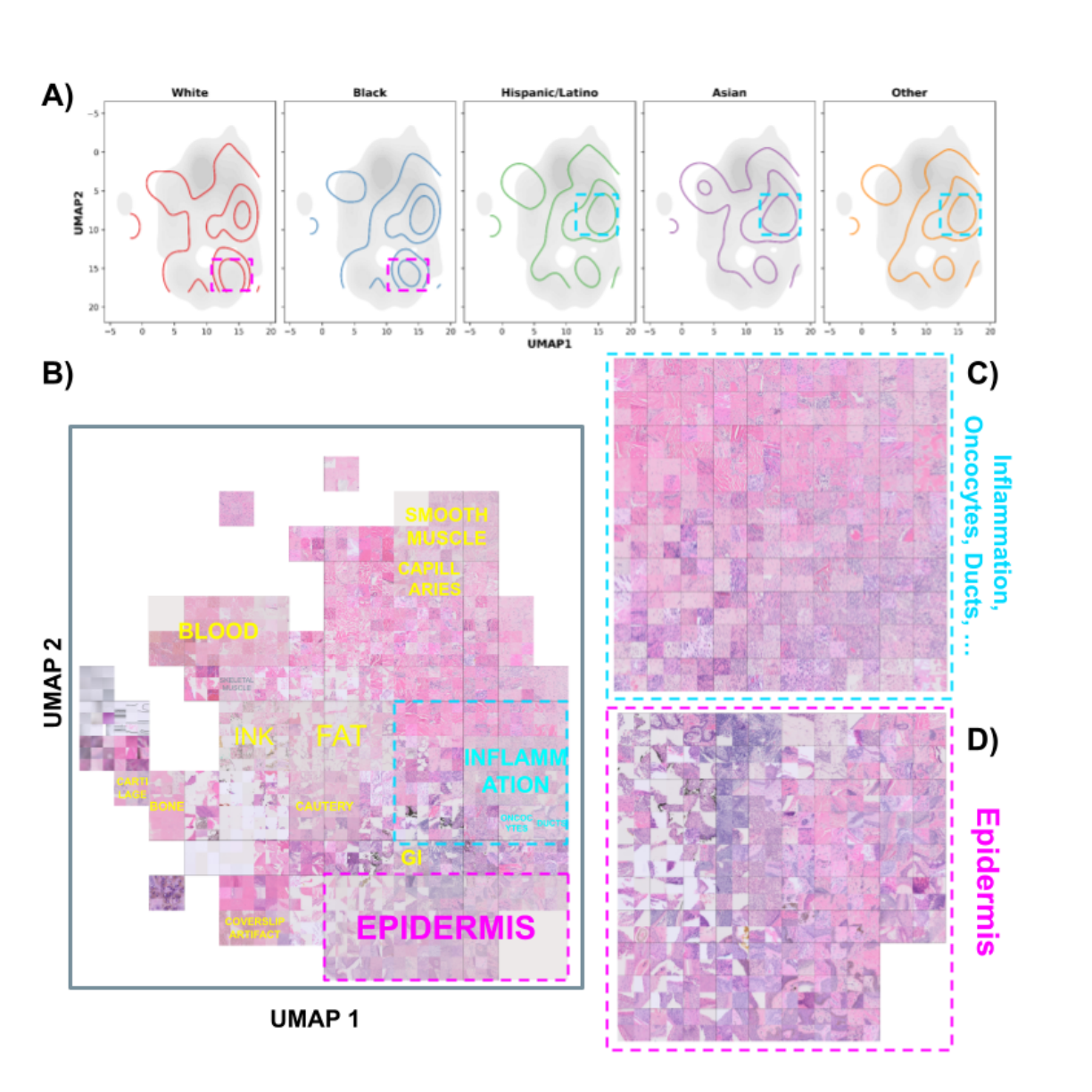 Figure 1: UMAP visualization of attention scores. (A) shows the high-attention regions (top 10%) for each racial group with contour lines, indicating that attention is concentrated in specific areas for White and Black groups. (B) through (D) show that high-attention regions are associated with specific tissue structures such as the "epidermis."
Figure 1: UMAP visualization of attention scores. (A) shows the high-attention regions (top 10%) for each racial group with contour lines, indicating that attention is concentrated in specific areas for White and Black groups. (B) through (D) show that high-attention regions are associated with specific tissue structures such as the "epidermis." -
Ablation Study: To confirm how crucial the epidermis was for prediction, an experiment was conducted where tiles from the epidermal region were intentionally removed from the validation data.
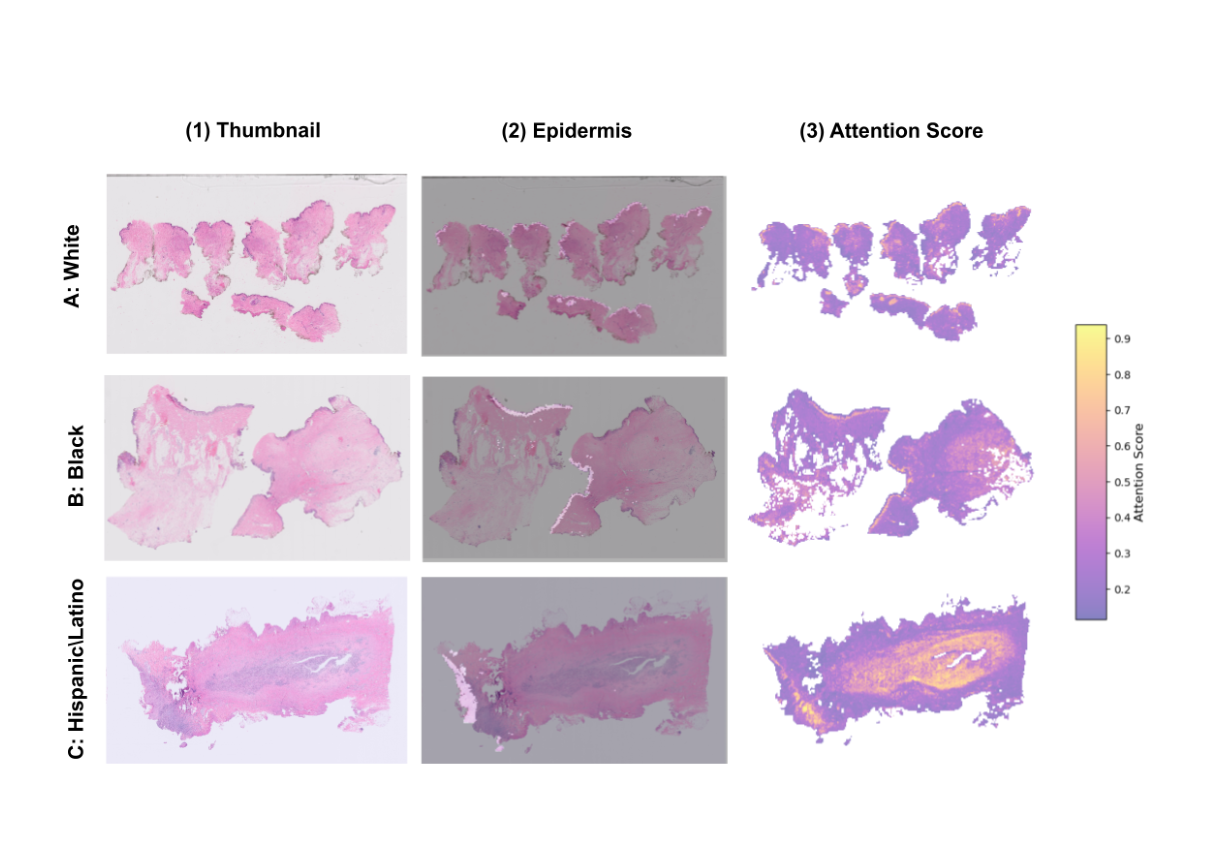 Figure 2: Attention and ablation analysis. (A) compares attention scores for epidermal and non-epidermal regions, showing that the epidermis receives higher attention in many groups. (B) shows the results of the ablation experiment, where removing epidermal tiles (orange) significantly drops performance from the original (green), and conversely, keeping only epidermal tiles (blue) maintains performance.
Figure 2: Attention and ablation analysis. (A) compares attention scores for epidermal and non-epidermal regions, showing that the epidermis receives higher attention in many groups. (B) shows the results of the ablation experiment, where removing epidermal tiles (orange) significantly drops performance from the original (green), and conversely, keeping only epidermal tiles (blue) maintains performance. -
The results confirmed that removing the epidermal region significantly degraded the model's performance, while conversely, performance was maintained even when only the epidermal region was left. This is definitive proof that the AI model uses the morphological features of the "epidermis" as a powerful shortcut for race prediction.
-
Applications and Future Prospects
Potential Applications
The findings of this research are not intended for direct application in products or services but are extremely important as "guidelines" for developing and evaluating medical AI.
- Integration into AI Development Processes: AI development companies need to incorporate bias analysis and interpretability methods, as shown in this study, into their development processes to ensure model fairness.
- Quality Assurance for Diagnostic AI: It is expected to be used as an auditing tool to verify that AI used in clinical settings does not depend on patient attributes unrelated to the disease.
Business Prospects
This research suggests new market opportunities for developing more reliable and fair AI.
- Development of Bias Detection/Mitigation Tools: Technologies and services that automatically detect hidden biases in AI models and mitigate their impact are likely to have high value in the future AI market.
- Trustworthy AI Consulting: The demand for specialized consulting services that assess the ethical and social risks of AI for medical institutions and development companies and propose countermeasures is expected to grow.
Future Challenges
While this study has provided important insights, several challenges remain.
- Correlation with Genetic Background: Since self-reported race also includes social aspects, future research needs to combine it with genetic ancestry data to analyze biological factors more precisely.
- Generalization to Other Organs: Further research is needed to determine if similar "shortcuts" exist in histology images of other organs (e.g., lung, colon).
- Development of Bias Mitigation Techniques: After identifying the shortcuts that cause bias, the next step is to develop specific bias mitigation techniques that guide the model not to rely on them.
Conclusion
This paper revealed that a deep learning model can predict a patient's self-reported race from skin histopathology images with moderate accuracy. It determined that this prediction is likely based on leveraging dataset biases, such as disease distribution, and using morphological features of tissues like the "epidermis" as a "shortcut."
These findings strongly suggest the necessity of carefully considering demographic biases when developing and evaluating AI models in computational pathology. To achieve fair and reliable medical AI, it is essential to constantly verify that the model is learning the intrinsic features of a disease and to make efforts to reduce the risk of relying on unintentional shortcuts.
Notes
- Note 1: Computational Pathology (CPath): A research field that supports cancer diagnosis, prognosis prediction, and more by analyzing digitized pathology tissue images with computers.
- Note 2: Shortcut Learning: A phenomenon where an AI model learns simple statistical correlations (shortcuts) accidentally present in the training data, rather than the essential rules for solving a task. This can cause a significant drop in performance on unseen data.
- Note 3: Attention-based Multiple Instance Learning (AB-MIL): A method that treats a large image (the whole slide) as a collection of small patches (tiles) and learns efficiently by paying "attention" to the important patches for prediction.
- Note 4: Confounding Factor: A third variable that is related to both the cause and the effect, creating a spurious relationship between them. In this study, a "specific disease" was related to both "race" and the "AI's prediction," acting as a factor that could lead to misinterpretation of causality.
- Note 5: Foundation Model (FM): A general-purpose AI model pre-trained on a very large dataset, such as text and images from the internet. By fine-tuning it for a specific task, high performance can be achieved.
- Note 6: UMAP (Uniform Manifold Approximation and Projection): A technique for compressing high-dimensional data into a lower dimension (usually 2D or 3D) while preserving its structure, for visualization purposes.
- Note 7: AUC (Area Under the Curve): A value representing the area under the ROC curve, used as a metric for evaluating the performance of classification models. A value closer to 1 indicates higher discriminative ability of the model.
Related Posts
2025-08-13
This paper presents a large-scale, systematic comparison and analysis of the performance of Transformer models, which have recently gained attention, and conventional mainstream CNN models for object detection tasks using remote sensing data such as satellite imagery. The study evaluates 11 different models on three datasets with distinct characteristics, revealing the potential for Transformers to outperform CNNs and clarifying the trade-off with the associated training costs.
2025-08-13
This paper proposes a new benchmark, 'Document Haystack,' which measures the ability to find specific information from long documents up to 200 pages long. This benchmark evaluates how accurately a Vision Language Model (VLM) can find intentionally embedded text or image information ('needles') within a document. The experimental results reveal that while current VLMs perform well on text-only documents, their performance significantly degrades on imaged documents or when handling information that combines text and images. This highlights future research challenges in the long-context and multimodal document understanding capabilities of VLMs.
2025-08-13
This paper proposes BF-PIP, a zero-shot method that utilizes Google's Gemini 2.5 Pro to predict pedestrian crossing intention without any additional training. Unlike conventional frame-based methods, it directly uses short, continuous videos and metadata such as ego-vehicle speed, achieving a high accuracy of 73% and demonstrating the potential for robust intention prediction based on contextual understanding.